The Renal Gene Expression Database (RGED) is the first resource for gene expression information from renal disease states. RGED stores and integrates different types of expression data and makes these data freely available in formats appropriate for comprehensive analysis. The initial version, RGED v1, was released on Oct 1st, 2013.
Gene Expression Plot
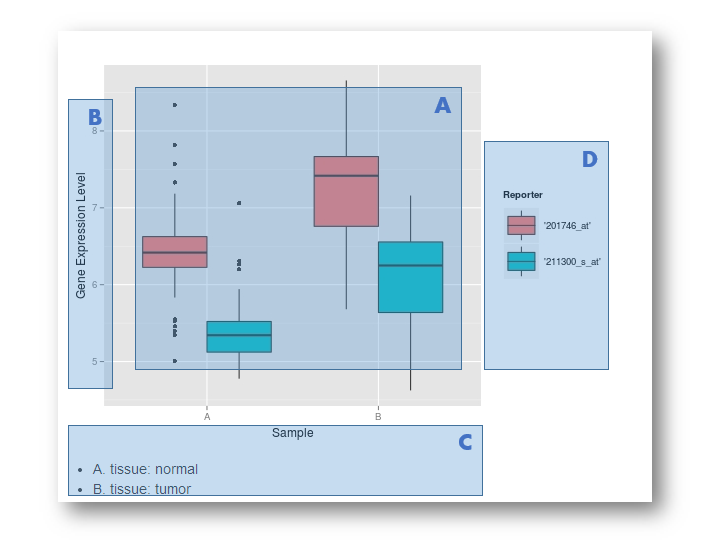
A. Box plot of gene expressions in the samples
B. Measurement of gene expression level
C. Descriptions of the samples used in the experiment
D. Each gene may have more than one reporter on different microarry designs, the expression level of each probe was measured individually.
Similarity Analysis
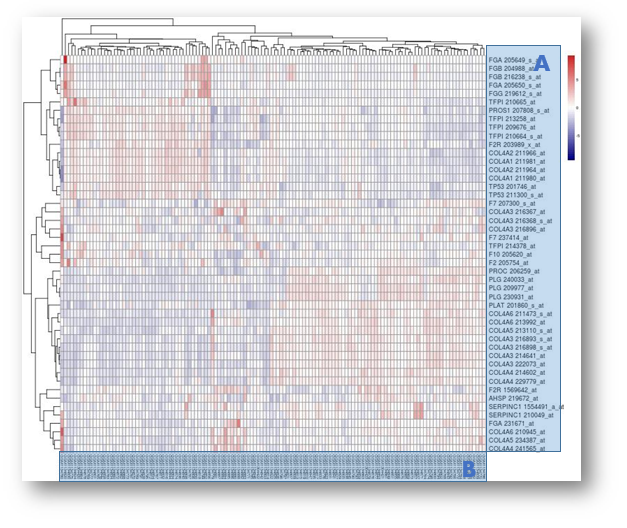
A. Genes of different reporters
B. Experimental samples
Using the Hierarchical Clustering algorithm, the gene of your interest can be searched against other gene sets, eg. Core Pathway Members. In this way, you can find out how similar it expressed with other gene candidates. Therefore, novel correlationship can be established among these genes.
News
- Aug 7, 2014
- Added RNA-seq experiment samples from the following experiments in Kidney Carcinoma research:
- Dec 12, 2013
- Added Samples from the following experiments in Chronic Kidney Disease and the Uremic Syndrom research:
- Added Samples from the following experiments in Acute Kidney Injury research:
- Oct 1, 2013
- RGED v1 is now open for public access! Search your gene, or browse the database by disease categories to find out more.