PIK3CA Expression Profiles in Kidney Transplantation
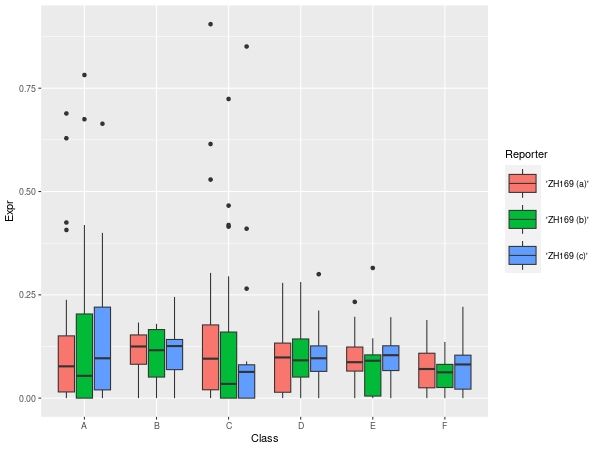
Tool Box: Similarity Analysis
Sample Grouping
| 1 | GSM456771 | AR1_Rep1 | condition_sample_id: AR1 | tissue type: kidney | state: acute rejection |
| 2 | GSM456772 | AR1_Rep2 | condition_sample_id: AR1 | tissue type: kidney | state: acute rejection |
| 3 | GSM456773 | AR2_Rep1 | condition_sample_id: AR2 | tissue type: kidney | state: acute rejection |
| 4 | GSM456774 | AR2_Rep2 | condition_sample_id: AR2 | tissue type: kidney | state: acute rejection |
| 5 | GSM456775 | AR3_Rep1 | condition_sample_id: AR3 | tissue type: kidney | state: acute rejection |
| 6 | GSM456776 | AR3_Rep2 | condition_sample_id: AR3 | tissue type: kidney | state: acute rejection |
| 7 | GSM456777 | AR4_Rep1 | condition_sample_id: AR4 | tissue type: kidney | state: acute rejection |
| 8 | GSM456778 | AR4_Rep2 | condition_sample_id: AR4 | tissue type: kidney | state: acute rejection |
| 9 | GSM456779 | AR5_Rep1 | condition_sample_id: AR5 | tissue type: kidney | state: acute rejection |
| 10 | GSM456780 | AR5_Rep2 | condition_sample_id: AR5 | tissue type: kidney | state: acute rejection |
| 11 | GSM456781 | AR6_Rep1 | condition_sample_id: AR6 | tissue type: kidney | state: acute rejection |
| 12 | GSM456782 | AR6_Rep2 | condition_sample_id: AR6 | tissue type: kidney | state: acute rejection |
| 13 | GSM456783 | AR7_Rep1 | condition_sample_id: AR7 | tissue type: kidney | state: acute rejection |
| 14 | GSM456784 | AR7_Rep2 | condition_sample_id: AR7 | tissue type: kidney | state: acute rejection |
| 15 | GSM456785 | AR8_Rep1 | condition_sample_id: AR8 | tissue type: kidney | state: acute rejection |
| 16 | GSM456786 | AR8_Rep2 | condition_sample_id: AR8 | tissue type: kidney | state: acute rejection |
| 17 | GSM456787 | AR9_Rep1 | condition_sample_id: AR9 | tissue type: kidney | state: acute rejection |
| 18 | GSM456788 | AR9_Rep2 | condition_sample_id: AR9 | tissue type: kidney | state: acute rejection |
| 19 | GSM456789 | AR10_Rep1 | condition_sample_id: AR10 | tissue type: kidney | state: acute rejection |
| 20 | GSM456791 | AR11_Rep1 | condition_sample_id: AR11 | tissue type: kidney | state: acute rejection |
| 21 | GSM456792 | AR11_Rep2 | condition_sample_id: AR11 | tissue type: kidney | state: acute rejection |
| 22 | GSM456793 | AR12_Rep1 | condition_sample_id: AR12 | tissue type: kidney | state: acute rejection |
| 23 | GSM456794 | AR12_Rep2 | condition_sample_id: AR12 | tissue type: kidney | state: acute rejection |
| 24 | GSM456795 | AR13_Rep1 | condition_sample_id: AR13 | tissue type: kidney | state: acute rejection |
| 25 | GSM456797 | AR14_Rep1 | condition_sample_id: AR14 | tissue type: kidney | state: acute rejection |
| 26 | GSM456798 | AR14_Rep2 | condition_sample_id: AR14 | tissue type: kidney | state: acute rejection |
| 27 | GSM456799 | AR15_Rep1 | condition_sample_id: AR15 | tissue type: kidney | state: acute rejection |
| 28 | GSM456800 | AR15_Rep2 | condition_sample_id: AR15 | tissue type: kidney | state: acute rejection |
| 1 | GSM456801 | ATN1_Rep1 | condition_sample_id: ATN1 | tissue type: kidney | state: acute tubular necrosis |
| 2 | GSM456802 | ATN1_Rep2 | condition_sample_id: ATN1 | tissue type: kidney | state: acute tubular necrosis |
| 3 | GSM456803 | ATN2_Rep1 | condition_sample_id: ATN2 | tissue type: kidney | state: acute tubular necrosis |
| 4 | GSM456804 | ATN2_Rep2 | condition_sample_id: ATN2 | tissue type: kidney | state: acute tubular necrosis |
| 5 | GSM456805 | ATN3_Rep1 | condition_sample_id: ATN3 | tissue type: kidney | state: acute tubular necrosis |
| 6 | GSM456806 | ATN3_Rep2 | condition_sample_id: ATN3 | tissue type: kidney | state: acute tubular necrosis |
| 7 | GSM456807 | ATN4_Rep1 | condition_sample_id: ATN4 | tissue type: kidney | state: acute tubular necrosis |
| 8 | GSM456809 | ATN5_Rep1 | condition_sample_id: ATN5 | tissue type: kidney | state: acute tubular necrosis |
| 9 | GSM456810 | ATN5_Rep2 | condition_sample_id: ATN5 | tissue type: kidney | state: acute tubular necrosis |
| 1 | GSM456811 | BL1_Rep1 | condition_sample_id: BL1 | tissue type: kidney | state: borderline change |
| 2 | GSM456812 | BL1_Rep2 | condition_sample_id: BL1 | tissue type: kidney | state: borderline change |
| 3 | GSM456813 | BL2_Rep1 | condition_sample_id: BL2 | tissue type: kidney | state: borderline change |
| 4 | GSM456814 | BL2_Rep2 | condition_sample_id: BL2 | tissue type: kidney | state: borderline change |
| 5 | GSM456815 | BL3_Rep1 | condition_sample_id: BL3 | tissue type: kidney | state: borderline change |
| 6 | GSM456816 | BL3_Rep2 | condition_sample_id: BL3 | tissue type: kidney | state: borderline change |
| 7 | GSM456817 | BL4_Rep1 | condition_sample_id: BL4 | tissue type: kidney | state: borderline change |
| 8 | GSM456818 | BL4_Rep2 | condition_sample_id: BL4 | tissue type: kidney | state: borderline change |
| 9 | GSM456819 | BL5_Rep1 | condition_sample_id: BL5 | tissue type: kidney | state: borderline change |
| 10 | GSM456820 | BL5_Rep2 | condition_sample_id: BL5 | tissue type: kidney | state: borderline change |
| 11 | GSM456821 | BL6_Rep1 | condition_sample_id: BL6 | tissue type: kidney | state: borderline change |
| 12 | GSM456822 | BL6_Rep2 | condition_sample_id: BL6 | tissue type: kidney | state: borderline change |
| 13 | GSM456823 | BL7_Rep1 | condition_sample_id: BL7 | tissue type: kidney | state: borderline change |
| 14 | GSM456824 | BL7_Rep2 | condition_sample_id: BL7 | tissue type: kidney | state: borderline change |
| 15 | GSM456825 | BL8_Rep1 | condition_sample_id: BL8 | tissue type: kidney | state: borderline change |
| 16 | GSM456827 | BL9_Rep1 | condition_sample_id: BL9 | tissue type: kidney | state: borderline change |
| 17 | GSM456828 | BL9_Rep2 | condition_sample_id: BL9 | tissue type: kidney | state: borderline change |
| 18 | GSM456829 | BL10_Rep1 | condition_sample_id: BL10 | tissue type: kidney | state: borderline change |
| 19 | GSM456830 | BL10_Rep2 | condition_sample_id: BL10 | tissue type: kidney | state: borderline change |
| 20 | GSM456831 | BL11_Rep1 | condition_sample_id: BL11 | tissue type: kidney | state: borderline change |
| 1 | GSM456833 | NR1_Rep1 | condition_sample_id: NR1 | tissue type: kidney | state: non-rejection |
| 2 | GSM456834 | NR1_Rep2 | condition_sample_id: NR1 | tissue type: kidney | state: non-rejection |
| 3 | GSM456835 | NR2_Rep1 | condition_sample_id: NR2 | tissue type: kidney | state: non-rejection |
| 4 | GSM456836 | NR2_Rep2 | condition_sample_id: NR2 | tissue type: kidney | state: non-rejection |
| 5 | GSM456837 | NR3_Rep1 | condition_sample_id: NR3 | tissue type: kidney | state: non-rejection |
| 6 | GSM456838 | NR3_Rep2 | condition_sample_id: NR3 | tissue type: kidney | state: non-rejection |
| 7 | GSM456839 | NR4_Rep1 | condition_sample_id: NR4 | tissue type: kidney | state: non-rejection |
| 8 | GSM456840 | NR4_Rep2 | condition_sample_id: NR4 | tissue type: kidney | state: non-rejection |
| 9 | GSM456841 | NR5_Rep1 | condition_sample_id: NR5 | tissue type: kidney | state: non-rejection |
| 10 | GSM456842 | NR5_Rep2 | condition_sample_id: NR5 | tissue type: kidney | state: non-rejection |
| 11 | GSM456843 | NR6_Rep1 | condition_sample_id: NR6 | tissue type: kidney | state: non-rejection |
| 12 | GSM456844 | NR6_Rep2 | condition_sample_id: NR6 | tissue type: kidney | state: non-rejection |
| 13 | GSM456845 | NR7_Rep1 | condition_sample_id: NR7 | tissue type: kidney | state: non-rejection |
| 14 | GSM456846 | NR7_Rep2 | condition_sample_id: NR7 | tissue type: kidney | state: non-rejection |
| 15 | GSM456847 | NR8_Rep1 | condition_sample_id: NR8 | tissue type: kidney | state: non-rejection |
| 16 | GSM456848 | NR8_Rep2 | condition_sample_id: NR8 | tissue type: kidney | state: non-rejection |
| 17 | GSM456849 | NR9_Rep1 | condition_sample_id: NR9 | tissue type: kidney | state: non-rejection |
| 18 | GSM456850 | NR9_Rep2 | condition_sample_id: NR9 | tissue type: kidney | state: non-rejection |
| 1 | GSM456851 | PR1_Rep1 | condition_sample_id: PR1 | tissue type: kidney | state: presumed rejection |
| 2 | GSM456852 | PR1_Rep2 | condition_sample_id: PR1 | tissue type: kidney | state: presumed rejection |
| 3 | GSM456853 | PR2_Rep1 | condition_sample_id: PR2 | tissue type: kidney | state: presumed rejection |
| 4 | GSM456854 | PR2_Rep2 | condition_sample_id: PR2 | tissue type: kidney | state: presumed rejection |
| 5 | GSM456855 | PR3_Rep1 | condition_sample_id: PR3 | tissue type: kidney | state: presumed rejection |
| 6 | GSM456856 | PR3_Rep2 | condition_sample_id: PR3 | tissue type: kidney | state: presumed rejection |
| 7 | GSM456857 | PR4_Rep1 | condition_sample_id: PR4 | tissue type: kidney | state: presumed rejection |
| 8 | GSM456858 | PR4_Rep2 | condition_sample_id: PR4 | tissue type: kidney | state: presumed rejection |
| 9 | GSM456859 | PR5_Rep1 | condition_sample_id: PR5 | tissue type: kidney | state: presumed rejection |
| 10 | GSM456860 | PR5_Rep2 | condition_sample_id: PR5 | tissue type: kidney | state: presumed rejection |
| 11 | GSM456861 | PR6_Rep1 | condition_sample_id: PR6 | tissue type: kidney | state: presumed rejection |
| 12 | GSM456862 | PR6_Rep2 | condition_sample_id: PR6 | tissue type: kidney | state: presumed rejection |
| 13 | GSM456863 | PR7_Rep1 | condition_sample_id: PR7 | tissue type: kidney | state: presumed rejection |
| 14 | GSM456864 | PR7_Rep2 | condition_sample_id: PR7 | tissue type: kidney | state: presumed rejection |
| 1 | GSM456865 | TX1_Rep1 | condition_sample_id: TX1 | tissue type: kidney | state: renal recipients with stable function |
| 2 | GSM456866 | TX1_Rep2 | condition_sample_id: TX1 | tissue type: kidney | state: renal recipients with stable function |
| 3 | GSM456867 | TX2_Rep1 | condition_sample_id: TX2 | tissue type: kidney | state: renal recipients with stable function |
| 4 | GSM456868 | TX2_Rep2 | condition_sample_id: TX2 | tissue type: kidney | state: renal recipients with stable function |
| 5 | GSM456869 | TX3_Rep1 | condition_sample_id: TX3 | tissue type: kidney | state: renal recipients with stable function |
| 6 | GSM456870 | TX3_Rep2 | condition_sample_id: TX3 | tissue type: kidney | state: renal recipients with stable function |
| 7 | GSM456871 | TX4_Rep1 | condition_sample_id: TX4 | tissue type: kidney | state: renal recipients with stable function |
| 8 | GSM456872 | TX4_Rep2 | condition_sample_id: TX4 | tissue type: kidney | state: renal recipients with stable function |
| 9 | GSM456873 | TX5_Rep1 | condition_sample_id: TX5 | tissue type: kidney | state: renal recipients with stable function |
| 10 | GSM456874 | TX5_Rep2 | condition_sample_id: TX5 | tissue type: kidney | state: renal recipients with stable function |
| 11 | GSM456875 | TX6_Rep1 | condition_sample_id: TX6 | tissue type: kidney | state: renal recipients with stable function |
| 12 | GSM456876 | TX6_Rep2 | condition_sample_id: TX6 | tissue type: kidney | state: renal recipients with stable function |
| 13 | GSM456877 | TX7_Rep1 | condition_sample_id: TX7 | tissue type: kidney | state: renal recipients with stable function |
| 14 | GSM456878 | TX7_Rep2 | condition_sample_id: TX7 | tissue type: kidney | state: renal recipients with stable function |
| 15 | GSM456879 | TX8_Rep1 | condition_sample_id: TX8 | tissue type: kidney | state: renal recipients with stable function |
| 16 | GSM456880 | TX8_Rep2 | condition_sample_id: TX8 | tissue type: kidney | state: renal recipients with stable function |
| 17 | GSM456881 | TX9_Rep1 | condition_sample_id: TX9 | tissue type: kidney | state: renal recipients with stable function |
| 18 | GSM456882 | TX9_Rep2 | condition_sample_id: TX9 | tissue type: kidney | state: renal recipients with stable function |
| 19 | GSM456883 | TX10_Rep1 | condition_sample_id: TX10 | tissue type: kidney | state: renal recipients with stable function |
| 20 | GSM456884 | TX10_Rep2 | condition_sample_id: TX10 | tissue type: kidney | state: renal recipients with stable function |
| 21 | GSM456885 | TX11_Rep1 | condition_sample_id: TX11 | tissue type: kidney | state: renal recipients with stable function |
| 22 | GSM456886 | TX11_Rep2 | condition_sample_id: TX11 | tissue type: kidney | state: renal recipients with stable function |
| 23 | GSM456887 | TX12_Rep1 | condition_sample_id: TX12 | tissue type: kidney | state: renal recipients with stable function |
| 24 | GSM456888 | TX12_Rep2 | condition_sample_id: TX12 | tissue type: kidney | state: renal recipients with stable function |
| 25 | GSM456889 | TX13_Rep1 | condition_sample_id: TX13 | tissue type: kidney | state: renal recipients with stable function |
| 26 | GSM456890 | TX13_Rep2 | condition_sample_id: TX13 | tissue type: kidney | state: renal recipients with stable function |
| 27 | GSM456891 | TX14_Rep1 | condition_sample_id: TX14 | tissue type: kidney | state: renal recipients with stable function |
| 28 | GSM456892 | TX14_Rep2 | condition_sample_id: TX14 | tissue type: kidney | state: renal recipients with stable function |
| 29 | GSM456893 | TX15_Rep1 | condition_sample_id: TX15 | tissue type: kidney | state: renal recipients with stable function |
| 30 | GSM456894 | TX15_Rep2 | condition_sample_id: TX15 | tissue type: kidney | state: renal recipients with stable function |
Gene Information
- Official Symbol
- PIK3CA
- Officical Full Name
- phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha
- Primary Source
- HGNC:8975
- See related
- MIM:171834 HPRD:01382
- Chromosome Location
- 3q26.3
- Gene Type
- protein-coding
- Also known as
- CLOVE CWS5 MCAP MCM MCMTC PI3K p110-alpha
- External Sources
- GUDMAP
Experiment Description
- Accession
- RGED19130
- Type
- Microarray
- Summary
- We aim to screen out different gene expression profile in renal recipients with stable function. Results from the various study components can help to diagnose renal allograft dysfunction with different causes by distinct gene expression profile. Keywords: acute rejection, acute tubular necrosis, borderline change, non-rejection, presumed rejection, renal recipients with stable function, Samples GSM456771-GSM456800: This study has been accomplished with 15 patients of acute rejection on the kidney.Technical replicates: 2 replicates(except AR10 and AR13) Samples GSM456801-GSM456810: This study has been accomplished with 5 patients of acute tubular necrosis on the kidney. Technical replicates: 2 replicates(except ATN4) Samples GSM456811-GSM456831: This study has been accomplished with 11 patients of borderline change on the kidney.Tecnical replicates:2 replicates(except BL8 and BL11) Samples GSM456833-GSM456850: This study has been accomplished with 9 patients of non-rejection on the kidney.Technical replicates: 2 replicates Samples GSM456851-GSM456864: This study has been accomplished with 7 patients of presumed rejection on the kidney.Technical replicates: 2 replicates Samples GSM456865-GSM456894: This study has been accomplished with 15 patients of renal recipients with stable function.Technical replicates: 2 replicates
- Experiment Platform
- Zhejiang university human 449 oligonucleotide array
- Data Source
- GSE19130